Generate A Plant Guild With Networkx
If you’ve heard of companion planting, plant guilds are the next logical extension of that idea. The plant guild that everyone has heard of is the Three Sisters (corn, beans, and squash). Let’s see what other plants might be beneficial.
First, start a GPT4 Code Interpreter chat.
Upload plants.csv, with the following remark:
plants.csv is a tab-separated file. Using networkx, build a directed graph.
The nodes are Common Name, and each node should be connected to other nodes
based on the comma-separated contents of Beneficial To.
Great! Let’s start by loading the data from the file you provided. We’ll take a look at the first few rows to understand its structure, and then we’ll proceed to build the directed graph using NetworkX.
# Importing necessary libraries import pandas as pd # Loading the tab-separated file file_path = '/mnt/data/plants_uniq.csv' data = pd.read_csv(file_path, sep='\t') # Showing the first few rows of the data data.head() RESULT Common Name Latin Name Beneficial To 0 Apple Malus domestica Chives, Dill, Garlic, Nasturtium 1 Artichoke Cynara cardunculus Mint, Tarragon, Catnip, Rue 2 Asparagus Asparagus officinalis Tomato, Parsley, Basil, Marjoram 3 Basil Ocimum basilicum Tomato, Pepper, Oregano, Chamomile 4 Beans Phaseolus vulgaris Corn, Sunflower, Cucumber, PotatoThe data consists of three columns:
- Common Name: The name of the plant.
- Latin Name: The Latin name of the plant.
- Beneficial To: A comma-separated list of other plants that the current plant is beneficial to.
We’ll use the “Common Name” as the nodes in the graph and create directed edges between nodes based on the “Beneficial To” column.
Let’s proceed to build the directed graph.
# Importing networkx import networkx as nx # Creating a directed graph G = nx.DiGraph() # Adding nodes and edges to the graph based on the data for index, row in data.iterrows(): common_name = row['Common Name'] beneficial_to = row['Beneficial To'].split(', ') G.add_node(common_name) for neighbor in beneficial_to: G.add_edge(common_name, neighbor) # Checking the number of nodes and edges in the graph num_nodes = G.number_of_nodes() num_edges = G.number_of_edges() num_nodes, num_edges RESULT (93, 278)The directed graph has been successfully created with 93 nodes and 278 edges. Each node represents a plant’s common name, and the directed edges connect the nodes based on the beneficial relationships between the plants.
Would you like to visualize the graph or perform any specific analysis on it?
Network visualization, please.
Sure! Visualizing a graph with 93 nodes and 278 edges can be a bit dense, so the graph might appear cluttered. However, we’ll do our best to represent it. Let’s proceed with the visualization.
# Importing necessary libraries for visualization import matplotlib.pyplot as plt # Setting up the plot size plt.figure(figsize=(12, 12)) # Drawing the graph pos = nx.spring_layout(G, seed=42) # Using spring layout for better visualization nx.draw(G, pos, with_labels=True, node_size=50, font_size=8, font_color="black", node_color="skyblue", edge_color="gray", arrowsize=10) # Displaying the plot plt.title("Directed Graph Representing Beneficial Relationships between Plants") plt.show()
Here is the visualization of the directed graph representing the beneficial relationships between plants. The nodes represent the common names of the plants, and the directed edges show the connections based on the “Beneficial To” relationships.
Due to the number of nodes and edges, the graph may appear dense. If you have any specific queries or need further analysis on particular nodes or subgraphs, please let me know!
Make a subgraph called 'Three Sisters', and add the nodes for Corn, Beans,
and Squash, as well as extending the graph out once based on what is in the
comma-separated Beneficial To field.
The “Three Sisters” subgraph will include the nodes for Corn, Beans, and Squash, as well as the nodes that are mentioned in the “Beneficial To” field for these three plants.
We’ll begin by identifying the nodes and edges related to Corn, Beans, and Squash and then extend the subgraph based on the “Beneficial To” field. Let’s start by constructing this subgraph.
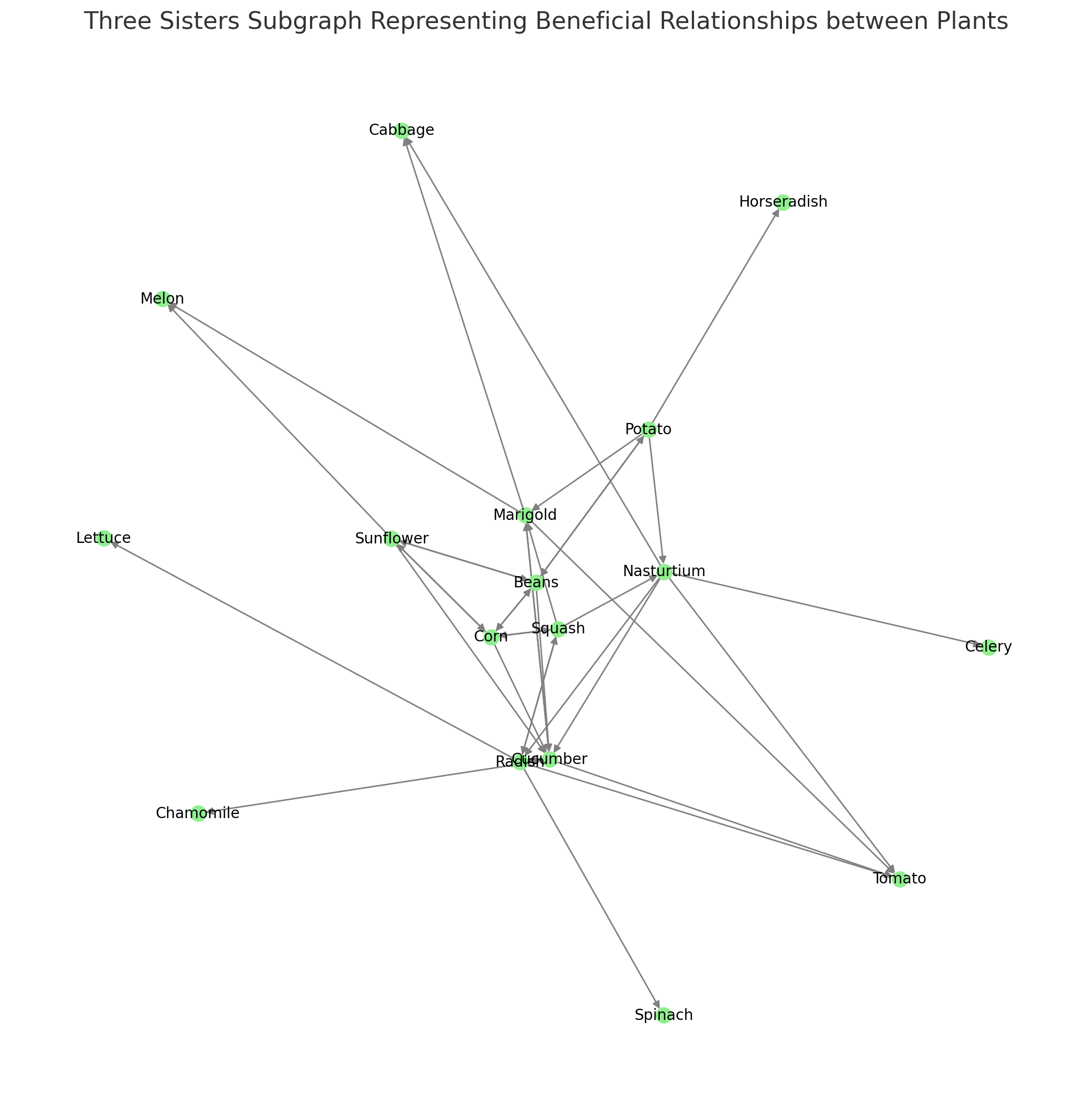
# Creating a subgraph called 'Three Sisters' with nodes for Corn, Beans, and Squash three_sisters_subgraph = nx.DiGraph() # Adding nodes and edges for Corn, Beans, and Squash three_sisters_plants = ["Corn", "Beans", "Squash"] for plant in three_sisters_plants: neighbors = list(G.successors(plant)) three_sisters_subgraph.add_node(plant) for neighbor in neighbors: three_sisters_subgraph.add_edge(plant, neighbor) # Extending the graph based on the 'Beneficial To' field (one level out) extended_neighbors = list(G.successors(neighbor)) three_sisters_subgraph.add_node(neighbor) for extended_neighbor in extended_neighbors: three_sisters_subgraph.add_edge(neighbor, extended_neighbor) # Checking the number of nodes and edges in the Three Sisters subgraph num_nodes_three_sisters = three_sisters_subgraph.number_of_nodes() num_edges_three_sisters = three_sisters_subgraph.number_of_edges() num_nodes_three_sisters, num_edges_three_sisters RESULT (17, 38)The “Three Sisters” subgraph has been successfully created with 17 nodes and 38 edges. It includes the nodes for Corn, Beans, and Squash, as well as the nodes that are connected to these three plants based on the “Beneficial To” field.
Would you like to visualize this subgraph as well?
Yes.
# Setting up the plot size plt.figure(figsize=(10, 10)) # Drawing the "Three Sisters" subgraph pos_three_sisters = nx.spring_layout(three_sisters_subgraph, seed=42) # Using spring layout for better visualization nx.draw(three_sisters_subgraph, pos_three_sisters, with_labels=True, node_size=100, font_size=10, font_color="black", node_color="lightgreen", edge_color="gray", arrowsize=10) # Displaying the plot plt.title("Three Sisters Subgraph Representing Beneficial Relationships between Plants") plt.show()
Here’s the visualization of the “Three Sisters” subgraph, representing the beneficial relationships between Corn, Beans, and Squash, as well as the plants connected to them through the “Beneficial To” field.
The nodes include the three main plants and their immediate neighbors, with the directed edges showing the connections.
If you have any more questions or need further assistance, please feel free to ask!